Admin Guide: Using Bulkrax CRC1280 Folder Parser To Do Imports
Using Bulkrax CRC1280 folder parser to do imports
Prepare the data
The Bulkrax CRC1280 folder parser expects the CRC1280 data to follow a very specific pattern, starting from a folder path within which is the group/experiment/subject/session/modality
The text file shows an example import_data tree import_data.txt
Copy the directory to be imported within CRC_FOLDER_IMPORT_PATH (the import data directory that is mounted onto the web and workers container as explained below), so it's available to the importer.
Note for developers / system administrators
-
The location of the test data folder in the host machine (from where you will be running docker) will need to be added to the .env file.
-
The environment variable is
CRC_FOLDER_IMPORT_PATH. -
This is mapped to the path
/rub-test-datain the container in docker (as defined in the docker-compose.yml)
ℹ After changing the contents of the .env file, stop the containers and start them again (updated values only apply after restarting the containers):
$ docker compose -f docker-compose.yml down
$ docker compose -f docker-compose.yml up -d
Import RUB s3 test data of CRC1280
-
rclone mount s3 bucket (ceph) with test data
rclone mount --daemon s3-rdms-test:fowi-rdms-testbucket/20220425_Test_GroupData /root/rdms.develop/hyrax/rub-test-data/20220425_Test_GroupData/⚠️ For reasons not yet fully understood, the
rclone mountprocess used to mount the S3 bucket into the bulk importer directory must be run asroot, even if the ReSeeD containers are started by a non-root user. We suspect that this is a limitation of the kernel'sfusemodule, and that therclone mountprocess must be run under the same UID as the importer processes inside the docker containers (which run as root, too).
Importing a subset of experiments from a file share
The CRC1280 importer will always import all data from /home/reseed/reseed/bulk-ingest (which is mounted as /rub-test-data inside the web and the worker containers). If you want to import only a subset of the experiments on a given fileshare, you will have to manually construct the directory structure that the CRC1280 importer expects for the experiments to be imported in /home/reseed/reseed/bulk-ingest, and then manually mount the corresponding directory for each experiment individually.
Steps to run an import
-
Log into ReSeeD as an administrator.
-
On the dashboard you should see the options Importers and Exporters. Click on Importers.

-
In the importers page, click on
Newon the top left corner. This would open the importer form
-
Fill in the Importer form

Name - Any name you would like to give the import job
Administrative set - choose the default admin set
Frequency - once
limit - empty
Parser - CRC1280 folder parser
Visibility - What you would like the visibility of the imported records to be.
Add folder path - Specify a path on the server
Import file path - /rub-test-data/test1/
Note:- The folder you want to import is test1
- test1 should contain the CRC1280 data starting from the group as shown in import_data.txt
- The data has been copied to the shared mount as stated in the section prepare the data).
Checking progress of the import
Importers page / dashboard
You can have an overview of the importer status from the Importers page

View page for each Importer
Clicking on the importer, would give you details on the current status of the import. You can also re-run an import from this page.
Import jobs are running

You should be able to monitor the status of the import of each job and view errors, if any.
Sidekiq interface to monitor background jobs and
You can also monitor background jobs running in Hyrax, including the background jobs created by the importer in the sidekiq interface.
Sidekiq is available at the endpoint /sidekiq (for example https://rdms.cottagelabs.com/sidekiq).
You need to logged as an administrator, to be able to view sidekiq.
From the interface you can monitor and administer all of the jobs in the queues and their status.
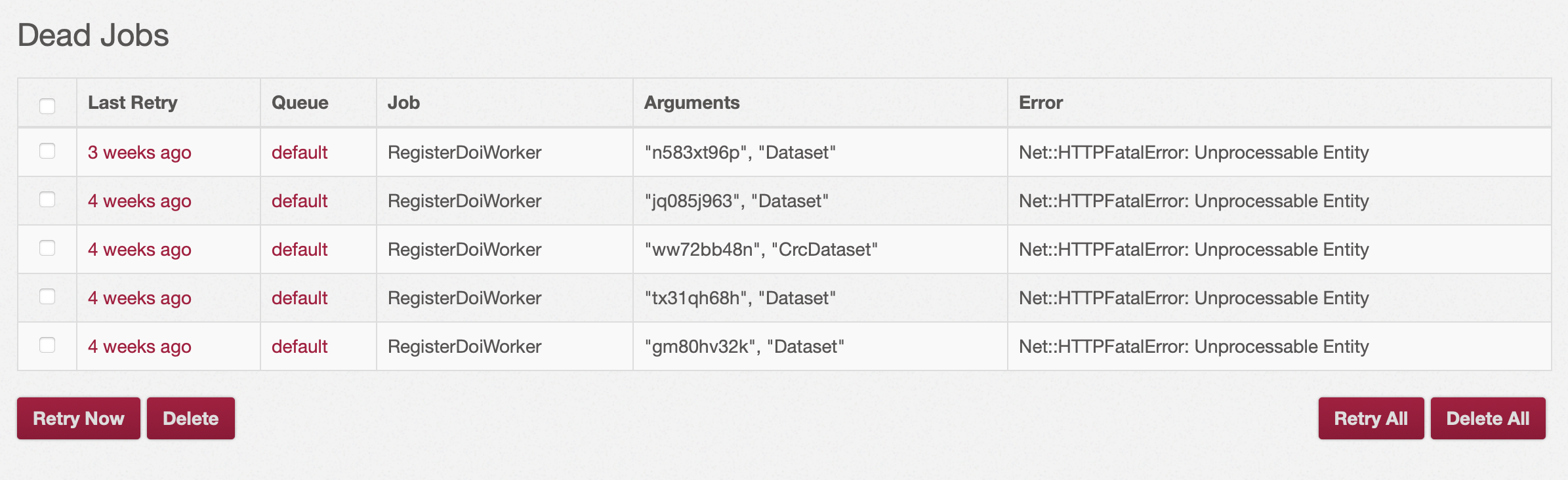
Note
There is one job you cannot monitor from the importer dashboard. It is the job scheduled to run after all of the collections, works and filesets have been imported. It is the job to create relationships between the collections, works and filesets. If this is not t=run, the uploaded files will not be associated to the filesets.

CSV file created by the importer
The importer first creates a csv file for the folder to be imported. This is then imported with a customised csv importer.
The csv files are stored at rdms/hyrax/tmp/imports/, where rdms is the root of the source directory checked out and from where the workers docker container is running.
Mark as completed and Rerun buttons
-
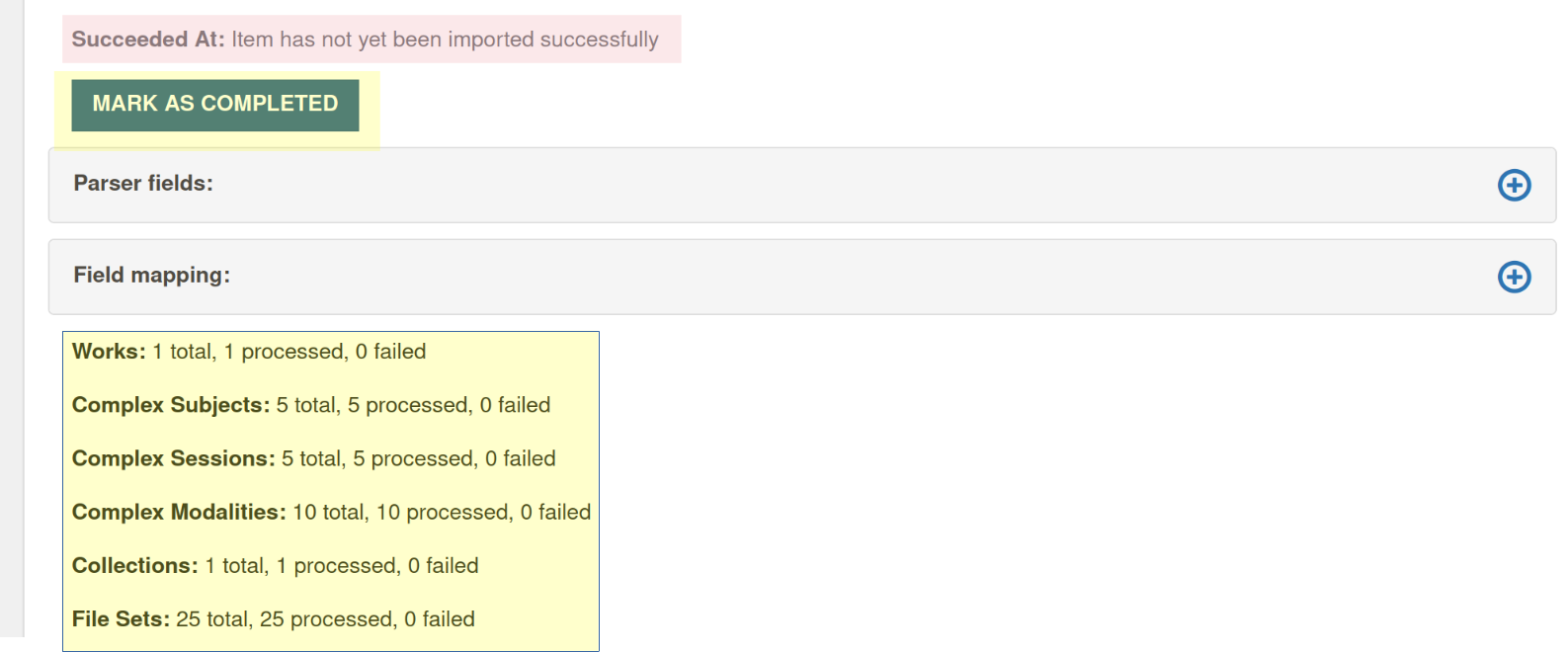
Mark as completed
This button is displayed the numbers show that the import has completed (total and number processed are the same, and the number failed is 0), but Bulkrax has got it's counting wrong and thinks the import is pending (like in the screen shot below). In such a case, it is safe to click on
Mark as completed. It will change the status of the import from pending to completed (and does nothing else).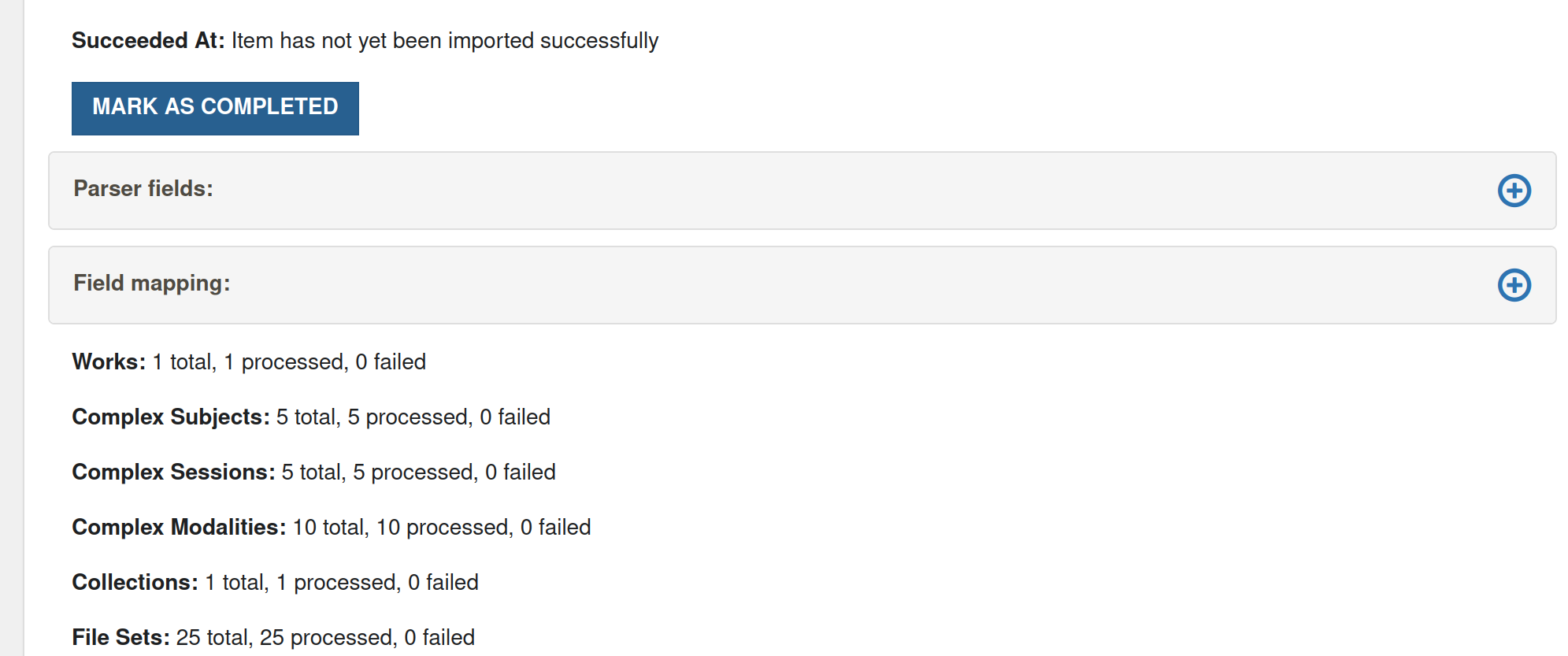
 `
` -
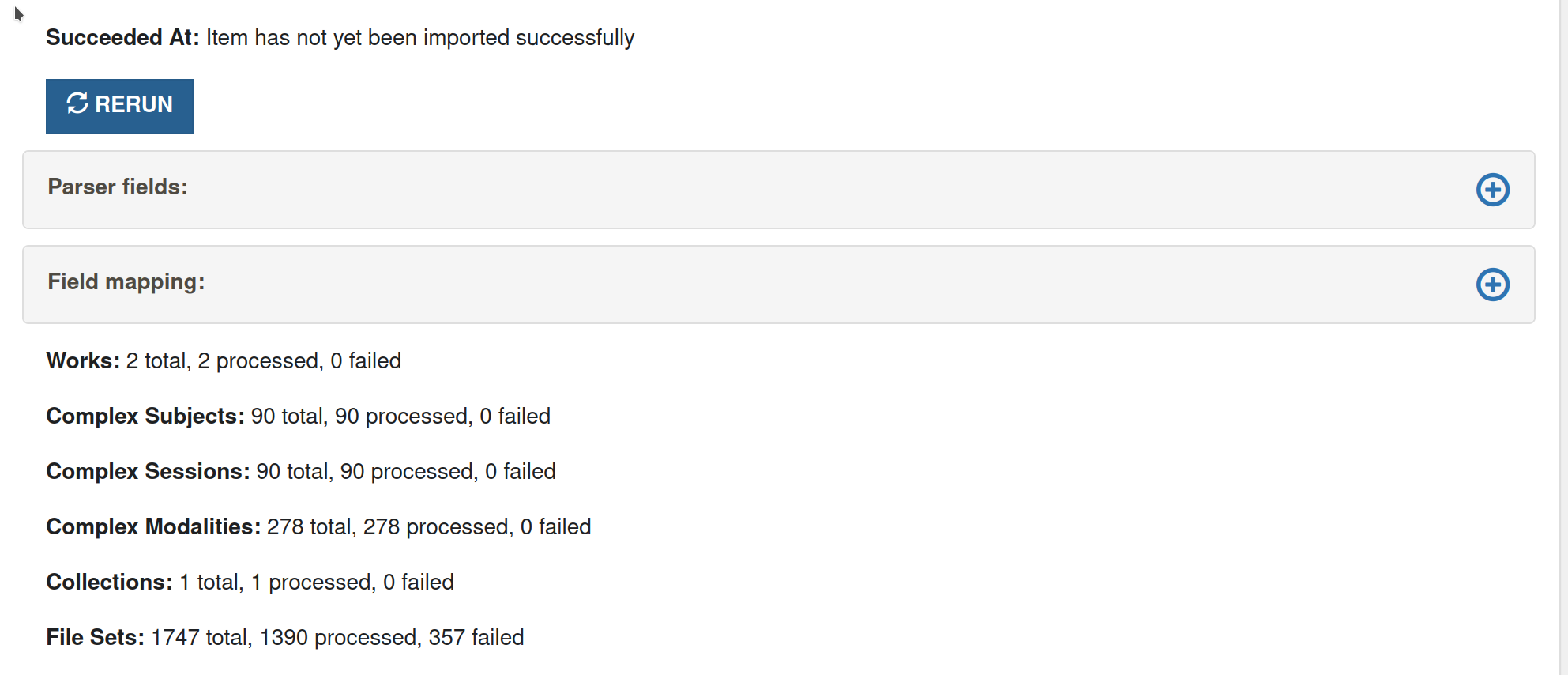
Rerun
This button is displayed when an import has completed, but with errors. When this button is clicked, it will cycle through all the entries in the importer and rerun all the failed jobs.
It is worth going into Sidekiq (for example - https://rdms.cottagelabs.com/sidekiq/. You need to logged in as admin to view this URL) first and checking if the job is still being processed in sidekiq (in busy, enqueued or retries). If it's an error like
Failed to acquire lock, then we have noticed that this goes away on a retry.If there are no jobs listed in sidekiq, you can click on rerun to rerun all the failed jobs.
Points to note
When a new CRCDataset is imported with the same collection name as an existing CRC1280 collection (either previously imported or created by a user), it will create a new collection, rather than reuse the one previously created.
When a new CRCDataset is imported again (it was previously imported), it will create a new collection and a new experiment, rather than reuse the ones previously created.
Troubleshooting
reschedule pending jobs from the rails console
- connect to the web container
docker exec -it web-1 /bin/bash rails c
importer = ::Bulkrax::Importer.find(importer_id)
# Needed only if the relationship job is missing.
# Adding it will do not harm.
# The job will run and n ot do anything if the relationships have finished.
::Bulkrax::ScheduleRelationshipsJob.set(wait: 5.minutes).perform_later(importer_id: importer.id)
importer.status_info('Pending')
importer.entries.each do |entry|
next if entry.status == 'Complete'
type = if entry.raw_metadata['model'] == 'CrcDataset'
'Work'
else
entry.raw_metadata['model']
end
entry.status_info('Pending')
"Bulkrax::CrcDataset::Import#{type}Job".constantize.send(
entry.parser.perform_method,
entry.id,
importer.importer_runs.last.id
)
end